Plugins¶
Here you can find a list of all the plugins that are currently available in your Laby instance.
Open Vector Editor (OVE)¶
OVE is a graphical sequence editor embedded in Laby that lets users view, design, edit, and annotate plasmids and DNA sequences. It provides synchronized circular and linear views, annotation and restriction-site handling, and sequence manipulation tools for common molecular biology workflows.
Integration with Laby¶
OVE can be embedded directly in Laby to link sequence data with inventory records and usage logs, improving plasmid management and traceability across the platform. Check a use case on the stocks product section See stocks_ove for details.
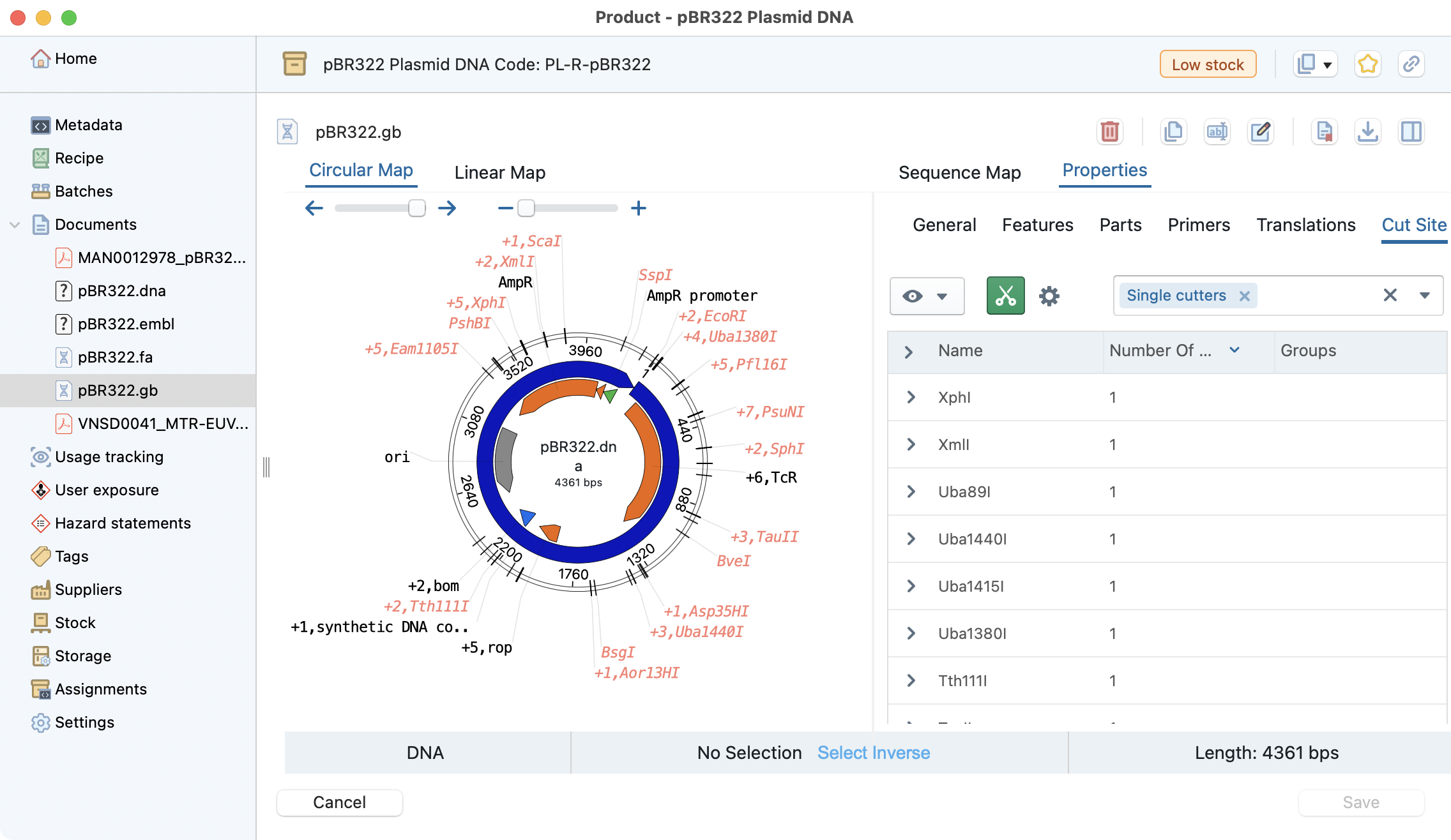
Common use cases¶
- Plasmid design and annotation
Enter or import sequences, add annotations (promoters, ORFs, markers, tags).
- Plasmid inventory management
Associate sequences with batches in Laby.
- In silico experiment simulation
Simulate digestions, PCRs, and other virtual workflows to predict fragments and outcomes.
Key features¶
Synchronized circular and linear views for concurrent selection and editing.
Annotation tools: create, edit, and import annotations (CSV, GenBank).
Auto-annotation to detect common genetic features.
Restriction digest simulation with fragment visualization and export.
Primer design and management utilities.
Protein translation from nucleotide sequences.
Support for common sequence formats and imports/exports.
Supported formats¶
Import/export examples: GenBank (.gb, .gbk), FASTA (.fasta), SnapGene (.dna), SBOL (.xml), and others.